 Genetics
- DNA & RNA Page
Genetics
- DNA & RNA Page  Genetics
- DNA & RNA Page
Genetics
- DNA & RNA Page
terms | concepts | structural components | functional process | argument | anomalies
In reality; genes and environment ecologically interact to form the world in which we thrive.
Chromosomes or "c’somes" must be understood from these two ways: structural versus functional.
basic terms | necessary concepts | chromosomes | functionality | epigenetic
Comprehending the terminology of the bio-microcosm
What turns the genes on and off?
Comprehending
the terminology of the biological-microcosm
or the world of the very small cells, molecules and compounds is a real challenge. The challenge arises from the complexity of proteins, amino acids, and chromosomes that act together to express and repress certain traits conveyed by genes.
Words:
Genes derive from the word generative or generate, meaning to give rise to something; as such they are the hereditary bequest.
generative - to give rise, to replicate or repair, reproductive ability.
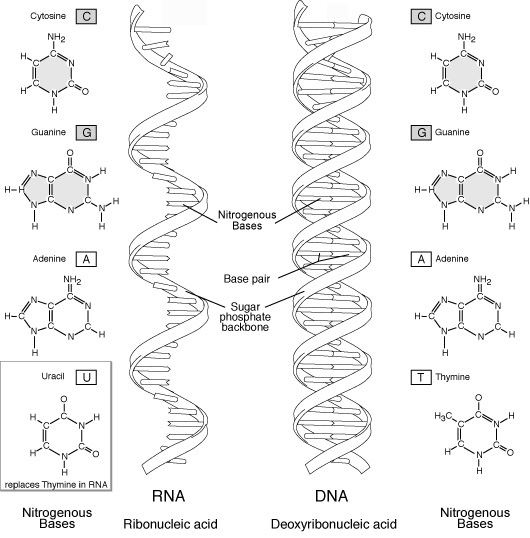
chromosome - elongated body of coherent nitrogen bases, helix shaped and coiled, found in the cell in more than one place: called chromosomes or C’somes.
allele - one of numerous forms of a gene, i.e. protein sequences in hemoglobin.
base pair - In DNA, a sequence of nitrogen compounds: A, T, G, C or U (RNA).
plasmid - small loops of encoding DNA residing in bacteria that replicate.
gene - the medium of inheritance: generally a trait, specifically a basic unit of heredity; some information (signal) capable of being inherited.
pangene - the original name given to cellular inheritance
gene pool - all the genes belonging to all the organisms in a population
gene frequency - For any population the rarity or commonness of one form of inheritable sequences (allele) rather than other forms on the same chromosome.
genetic drift - Changes over time in the genetic composition of populations due strictly to chance or statistical variation; often greater in small populations.
genome - the entire component of any organism’s genetic material.
• genotype - the molecularly encoded material make-up of C’somes.
genotype refers to what actually is hidden in the base pair sequences - codons
• phenotype - the observed expression of the underlying DNA sequences.phenotype refers to what is seen, sensed, encountered, smelled or heard, the expression o of the code in the form of proteins.
terminology | concepts | structural components | functional process | argument | anomalies
Concepts
antibiotic resistance - survival capacity of bacteria to withstand assault.
speciation - a process where geographical or other (behavioral) barriers arise
to separate organisms into two different and distinct breeding populations.
Argument is classical in its beginnings and asks -- does our inheritance or the environment determine the characteristics we see in creatures and ourselves?
nature (genetics) versus nurture (environment) as reasons for behavior.
adaptation as fitness; adjusting to ecological change is confused with the capacity to leave offspring that survive into an adult or reproductive stage.
Dialectical
materialism - belief
that existence is binary and best understood as a unity of
opposite tendencies. In the struggle of opposites a driving force propels everything
on.
technological determinism - a set ideals or an ideology which expresses a belief that existence is largely governed by how tools and technical systems force people to react.
Words II
generative - to give rise, to replicate or repair, reproductive ability.
The regenerative ability of all life, that is the capability of an organism
to exploit available niches, has been studied from both a structural and a functional
perspective.
Structural refers to how the component parts come together to form a larger whole. Whereas the functional view looks at how the process of bringing different parts together enables ingredients to become assembled into some opportunity, product or outcome.
Chromosomes or c’somes must be understood
from these two ways:
DNA or deoxyribonucleic acid is the material passed on from one generation to another by means of either mitotic (simple) division, or meiotic (reduction) division of cells or parts of cells [such as: the chloroplast and mitochondria).
Codon - section of base pairs that --in sets of three-- determine amino
acids.
introns - sections of the base pairs that intrude, with no apparent sequence into the codon and do not instruct the formation of amino acids
exons - those stretches of the codon that instruct the RNA to form one in particular of the twenty amino acids
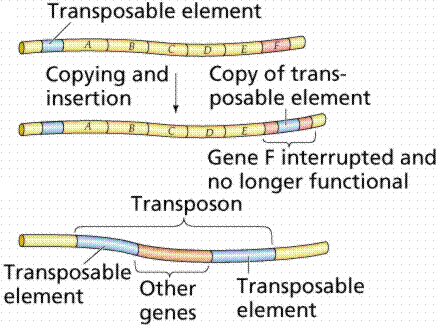
transposons - those exons sections of the codons that appear to move from one chromosome to another, for reasons, as yet, not well understood.
junk DNA - significant amounts of DNA that do not contribute instructions for making any of the sixty to eighty thousand genes, for instance, that humans possess.
chromosomes contain the DNA material that RNA edits in order to build amino
acid sequences in the ribosomes into proteins
base pairs - are DNA composed of four nitrogen bases: adenine, guanine, cytosine
and thymine which are derived from inorganic sources in the environment. For
example UUU (three uracil bases) codes for phenylalanine and amino acid.
amino acids - 20 varieties of macromolecule whose sequence determine the kind
of protein created in the ribosomes by RNA in cells, or in organelles.
proteins -are builders, nourishes, and decomposers of life in that they form
enzymes that change, and hormones that regulate, and lipoprotein structures
that maintain metabolic, homeostasis and reproductive activities in all living
things.
protein folding - refers to the means by which an inert sequence of amino acids
(which form the protein and whose sequences are dictated by the DNA- base pairs--)
are designed into a functional unit that can perform tasks.
2). Functional approach
“The fundamental evolutionary event is a change in the frequency of genes
and chromosome configurations in a population.” (over time) Hence over
several generations.”
Edward O. Wilson, The Diversity of Life: TDOL, p. 75.
Enzymes are functioning proteins that bring about some desired state of conditions when a preceding state triggers their creation by RNA, DNA and protein synthesis in the endoplasmic reticulum of the cell. Enzymes are proteins that bring about a change–catalyze reactions–needed for living responses to altering conditions.
“The human body contains approximately one trillion cells (one million
squared).... Inside the cell there is...the nucleus.....Inside the nucleus are
two complete sets of the human genome.... each set includes the same 60,000
to 80,000 genes on the same twenty-three chromosomes.”
“The idea of the genome as a book is not, strictly speaking, even a metaphor.
It is literally true. A book is a piece of digital information, written in linear,
one dimensional and one-directional form. . . .So is the genome.”
Matt Ridley, pp. 6-7.
Counterpoint:
"This is the metaphor of the empty bucket. Genes determine the size of the bucket and the environment determines how much is poured into it. If the environment is poor, then none of the buckets will have much in it at all and all genotypes will do poorly."“The organism is determined neither by its genes nor by its environment nor even by the interaction between them, but bears a significant mark of random processes."
Richard Lewontin, The Triple Helix. p. 38.
“There is a family of genes called apolipoprotein genes, or APO genes. They come in four basic varieties…though there are various different versions on of each on different chromosomes.”
“The job of APOEs protein is to…effect an introduction between VLDL
and a receptor on a cell that needs some triglycerides.”
Matt Ridley,
p. 259.
contents:
terminology | concepts | structural components | functional process | argument | anomalies
Scientific American, March 2007.
The epigenetic conundrum ![]() , in what direction does shifting occur?
, in what direction does shifting occur?
 Discoveries in recent years made it increasingly clear that there is far more to genetics than the sequence of building blocks that make up our genes. Adding molecules such as methyl groups to the backbone of DNA without altering the letters of the DNA alphabet can change how genes interact with the cell's transcribing machinery and hand cells an additional tool to fine-tune gene expression.
Discoveries in recent years made it increasingly clear that there is far more to genetics than the sequence of building blocks that make up our genes. Adding molecules such as methyl groups to the backbone of DNA without altering the letters of the DNA alphabet can change how genes interact with the cell's transcribing machinery and hand cells an additional tool to fine-tune gene expression.
"The goal of our study was to integrate multiple levels of epigenetic information since we still have a very poor understanding of the genome-wide regulation of methylation and its effect on the transcriptome," explains postdoctoral researcher and co-first author Ryan Lister, Ph.D.
The transcriptome encompasses all RNA copies or transcripts made from DNA. The bulk of transcripts consists of messenger RNAs, or mRNAs, that serve as templates for the manufacture of proteins but also includes regulatory small RNAs, or smRNAs. The latter wield their power over gene expression by literally cutting short the lives of mRNAs or tagging specific sequences in the genome for methylation.
Cells employ a whole army of enzymes that add methyl groups at specific sites, maintain established patterns or remove undesirable methyl groups. When Lister and his colleagues compared normal cells with cells lacking different combination of enzymes they discovered that cells put a lot of effort in keeping certain areas of the genome methylation-free.
On the flip side, the Salk researchers found that when they knocked out a whole class of methylases, a different type of methylase would step into the breach for the missing ones. This finding is relevant for a new class of cancer drugs that work by changing the methylation pattern in tumor cells.
"You might succeed in removing one type of methylation but end up with increasing a different type," says Ecker. "But very soon we will be able to look and see what kind of compensatory changes are happening and avoid unintended consequences."
Previous studies had found that a subset of smRNAs could direct methylation enzymes to the region of genomic DNA to which they aligned. Overlaying genome-wide methylome and smRNA datasets confirmed increased methylation precisely within the stretch of DNA that matched the sequence of the smRNA. Conversely, heavily methylated smRNA loci tended to spawn more smRNAs.
The New Scientist reported in April 2008,
If the genome is like a list of genetic ingredients, then the rules for how those genes are used and when they are switched on and off is the business of epigenetics. The first full piece of this "cookbook" has now been sequenced – a plant's epigenome.
Life often modifies its genetic material without changing the letters of the genetic code. One of the main ways this is done is through the addition of a chemical unit called a methyl group to a gene.
This methylation effectively gums up a gene's copying machinery. It is thought to be an important factor in directing stem cells to develop into different tissues, and problems with methylation are implicated in a number of diseases including cancer and Huntington's.
"Complete 'cookbook' for running a genome published," New Scientist,14:00 21 April 2008
* NewScientist.com news service
WHAT THIS AND RECENT FINDINGS MEAN IS THAT:
"This discovery broadens our view of methyltransferases and tells us that epigenetic regulation in cells is even more complicated than we thought," says principal investigator, Xiaodong Cheng, Ph.D., professor of biochemistry at Emory University School of Medicine. " This is an important piece of the puzzle, and additional research will continue to help us unwind the multiple mechanisms involved in epigenetic gene regulation."
contents:
terminology | concepts | structural components | functional process | argument | anomalies
![]() Keller | Lewontin | Margulis | Thomas
Keller | Lewontin | Margulis | Thomas
Genetics Index | What makes genetics significant? | History of Genetics | DNA discovery | RNA | Resistance| | Visual images